Thiessen polygon, also called Voronoi diagram, is a partition of a plane into regions close to each of a given set of objects. Thiessen polygons are be constructed around each sampled point so all the space within a specific polygon is closest in distance to that sampled point (comparing to other sampled points).
scipy package offers Voronoi() function to create Thiessen polygons.
Data Preparation
National Land Survey of Finland (https://www.maanmittauslaitos.fi/en) provides the latest shapefile for the Finnish administration boarders, which includes the municipalities.
OpenCellID (https://opencellid.org/) is a collaborative community project that collects GPS positions of cell towers and their corresponding location area identity.
In this section, we will map the collected cell towers to each municipalities and provinces, and export the merged dataframe.
When download the dataset which contains the cell towers’ GPS information, it is in EPSG:4326 coded. While the shapefile for Finnish municipality and province, the CRS is EPSG:3067.
import pandas as pd
import geopandas as gpd
import matplotlib.pyplot as plt
import contextily as ctx
from geopandas import GeoDataFrame
from shapely.geometry import Point
import pyproj
import warnings
warnings.filterwarnings('ignore')# load DNA LTE cell towers list downloaded from OpenCellID
dna_cell_tower = pd.read_csv('dna_lte.csv')
dna_cell_tower.head()| Unnamed: 0 | MCC | MCNC | eNB_ID | Longitude | Latitude | |
|---|---|---|---|---|---|---|
| 0 | 7489 | 244 | 12 | 2 | 24.607037 | 60.731043 |
| 1 | 7490 | 244 | 12 | 3 | 24.400345 | 60.997403 |
| 2 | 7491 | 244 | 12 | 4 | 24.613302 | 60.706910 |
| 3 | 7492 | 244 | 12 | 5 | 24.499218 | 60.912731 |
| 4 | 7493 | 244 | 12 | 6 | 24.510986 | 60.884484 |
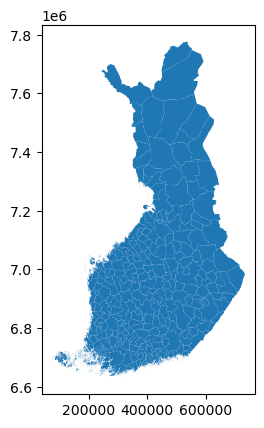
# load Finland municipality boundary spatial data downloaded from National Land Survey of Finland
finland_municipality = gpd.GeoDataFrame.from_file('kunta1000k_2023Polygon.shp')
finland_municipality.plot()<Axes: >
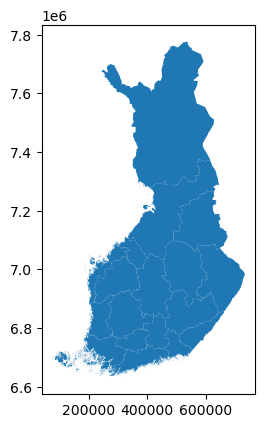
# load Finland province boundary spatial data downloaded from National Land Survey of Finland
finland_province = gpd.GeoDataFrame.from_file('maakunta1000k_2023Polygon.shp')
finland_province.plot()<Axes: >
# create a function to map each municipality to province
def municipality2province(municipality, province):
for ind in province.index:
if province['geometry'][ind].contains(municipality):
return province['name'][ind]
finland_municipality['province'] = finland_municipality['geometry'].apply(lambda x: municipality2province(x, finland_province))
finland_municipality.head()| kunta | vuosi | nimi | namn | name | geometry | province | |
|---|---|---|---|---|---|---|---|
| 0 | 005 | 2023 | Alajärvi | Alajärvi | Alajärvi | POLYGON ((366787.924 7001300.583, 364487.590 6... | South Ostrobothnia |
| 1 | 009 | 2023 | Alavieska | Alavieska | Alavieska | POLYGON ((382543.364 7120022.976, 382899.505 7... | North Ostrobothnia |
| 2 | 010 | 2023 | Alavus | Alavo | Alavus | POLYGON ((343298.204 6961570.195, 343831.847 6... | South Ostrobothnia |
| 3 | 016 | 2023 | Asikkala | Asikkala | Asikkala | POLYGON ((436139.680 6798279.085, 435714.468 6... | Päijät-Häme |
| 4 | 018 | 2023 | Askola | Askola | Askola | POLYGON ((426631.036 6720528.076, 428821.749 6... | Uusimaa |
Data Wrangling
In the following we will transform the GPS (WGS84 or EPSG:4326) to EPSG3067, and then merge two datasets and count the number of cell towers per municipality.
# create several functions to transform the projections
wgs84 = pyproj.Proj(projparams = 'epsg:4326')
finProj = pyproj.Proj(projparams = 'epsg:3067')
def crs_transformer(x, y):
return pyproj.transform(wgs84,finProj, y, x)
def transformed_x(x,y):
return crs_transformer(x,y)[0]
def transformed_y(x,y):
return crs_transformer(x,y)[1]# transform the cell tower's coordinates to EPSG3067
dna_cell_tower['Proj_Lon'] = dna_cell_tower.apply(lambda x: transformed_x(x['Longitude'], x['Latitude']), axis=1)
dna_cell_tower['Proj_Lat'] = dna_cell_tower.apply(lambda x: transformed_y(x['Longitude'], x['Latitude']), axis=1)
dna_cell_tower.head()| Unnamed: 0 | MCC | MCNC | eNB_ID | Longitude | Latitude | Proj_Lon | Proj_Lat | |
|---|---|---|---|---|---|---|---|---|
| 0 | 7489 | 244 | 12 | 2 | 24.607037 | 60.731043 | 369501.553119 | 6.735208e+06 |
| 1 | 7490 | 244 | 12 | 3 | 24.400345 | 60.997403 | 359409.547832 | 6.765288e+06 |
| 2 | 7491 | 244 | 12 | 4 | 24.613302 | 60.706910 | 369745.411927 | 6.732509e+06 |
| 3 | 7492 | 244 | 12 | 5 | 24.499218 | 60.912731 | 364394.910111 | 6.755654e+06 |
| 4 | 7493 | 244 | 12 | 6 | 24.510986 | 60.884484 | 364913.374089 | 6.752485e+06 |
# create a GeoDataFrame with each cell location as a Point
dna_cell_tower_gdf = gpd.GeoDataFrame(dna_cell_tower, geometry=gpd.points_from_xy(dna_cell_tower['Proj_Lon'], dna_cell_tower['Proj_Lat'], crs='EPSG:3067'))
dna_cell_tower_gdf.head()| Unnamed: 0 | MCC | MCNC | eNB_ID | Longitude | Latitude | Proj_Lon | Proj_Lat | geometry | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 7489 | 244 | 12 | 2 | 24.607037 | 60.731043 | 369501.553119 | 6.735208e+06 | POINT (369501.553 6735208.061) |
| 1 | 7490 | 244 | 12 | 3 | 24.400345 | 60.997403 | 359409.547832 | 6.765288e+06 | POINT (359409.548 6765288.274) |
| 2 | 7491 | 244 | 12 | 4 | 24.613302 | 60.706910 | 369745.411927 | 6.732509e+06 | POINT (369745.412 6732508.849) |
| 3 | 7492 | 244 | 12 | 5 | 24.499218 | 60.912731 | 364394.910111 | 6.755654e+06 | POINT (364394.910 6755653.753) |
| 4 | 7493 | 244 | 12 | 6 | 24.510986 | 60.884484 | 364913.374089 | 6.752485e+06 | POINT (364913.374 6752484.785) |
# merge cell tower dataframe and municipality dataframe
merge_gdf = dna_cell_tower_gdf.sjoin(finland_municipality[['kunta','name','geometry','province']], how='left')
merge_gdf.head()| Unnamed: 0 | MCC | MCNC | eNB_ID | Longitude | Latitude | Proj_Lon | Proj_Lat | geometry | index_right | kunta | name | province | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 7489 | 244 | 12 | 2 | 24.607037 | 60.731043 | 369501.553119 | 6.735208e+06 | POINT (369501.553 6735208.061) | 143.0 | 433 | Loppi | Kanta-Häme |
| 1 | 7490 | 244 | 12 | 3 | 24.400345 | 60.997403 | 359409.547832 | 6.765288e+06 | POINT (359409.548 6765288.274) | 41.0 | 109 | Hämeenlinna | Kanta-Häme |
| 2 | 7491 | 244 | 12 | 4 | 24.613302 | 60.706910 | 369745.411927 | 6.732509e+06 | POINT (369745.412 6732508.849) | 143.0 | 433 | Loppi | Kanta-Häme |
| 3 | 7492 | 244 | 12 | 5 | 24.499218 | 60.912731 | 364394.910111 | 6.755654e+06 | POINT (364394.910 6755653.753) | 54.0 | 165 | Janakkala | Kanta-Häme |
| 4 | 7493 | 244 | 12 | 6 | 24.510986 | 60.884484 | 364913.374089 | 6.752485e+06 | POINT (364913.374 6752484.785) | 54.0 | 165 | Janakkala | Kanta-Häme |
# save the merged dataframe
merge_gdf.to_csv('DNA_LTE_location.csv', index=False)# select Uusimaa area (where Helsinki is) for the analysis
uusimaa_cell = merge_gdf[merge_gdf['province'] == 'Uusimaa']
uusimaa_municipality = finland_municipality[finland_municipality['province'] == 'Uusimaa']
uusimaa_municipality.head()| kunta | vuosi | nimi | namn | name | geometry | province | |
|---|---|---|---|---|---|---|---|
| 4 | 018 | 2023 | Askola | Askola | Askola | POLYGON ((426631.036 6720528.076, 428821.749 6... | Uusimaa |
| 11 | 049 | 2023 | Espoo | Esbo | Espoo | MULTIPOLYGON (((372736.667 6661003.115, 372854... | Uusimaa |
| 26 | 078 | 2023 | Hanko | Hangö | Hanko | MULTIPOLYGON (((272609.681 6632304.439, 272418... | Uusimaa |
| 32 | 091 | 2023 | Helsinki | Helsingfors | Helsinki | MULTIPOLYGON (((379681.453 6664563.666, 379459... | Uusimaa |
| 33 | 092 | 2023 | Vantaa | Vanda | Vantaa | POLYGON ((393916.943 6694490.093, 394375.772 6... | Uusimaa |
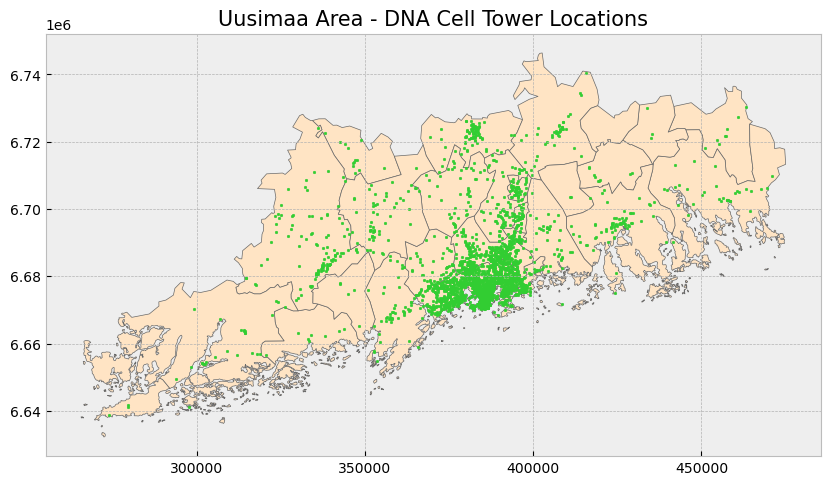
# Create subplots
fig, ax = plt.subplots(1, 1, figsize = (10, 10))
# Stylize plots
plt.style.use('bmh')
# Plot data
uusimaa_municipality.plot(ax = ax, color = 'bisque', edgecolor = 'dimgray')
uusimaa.plot(ax = ax, marker = 'o', color = 'limegreen', markersize = 3)
# Set title
ax.set_title('Uusimaa Area - DNA Cell Tower Locations', fontdict = {'fontsize': '15', 'fontweight' : '3'})Text(0.5, 1.0, 'Uusimaa Area - DNA Cell Tower Locations')
Thiessen Polygons
When creating Thiessen polygons, the sample points toward the edges of the point shapefile’s extent will have infinite Voronoi regions, because not all sides of these edge points have adjacent sample points that would constrain the regions. Consequently, these infinite regions will not be exported. To mitigate this issue, we can create dummy points well beyond the extent of our datasets, which will create finite Voronoi regions for all of our actual sample points. Then, we can clip the regions to our extent shapefile (creating dummy points far away from our actual sample points will ensure the dummy points and their infinite Voronoi regions do not interfere with the sample points and their associated finite Voronoi regions after all regions are clipped).
from scipy.spatial import Voronoi, voronoi_plot_2d
from shapely.geometry import Polygon
# Get X and Y coordinates of cell towers
x = uusimaa_cell["geometry"].x
y = uusimaa_cell["geometry"].y
# Create list of XY coordinate pairs
coords = [list(xy) for xy in zip(x, y)]# extend extent of municipality feature by using buffer
uusimaa_municipality_buffer = uusimaa_municipality.buffer(100000)
# get extent of buffered input feature
min_x_tp, min_y_tp, max_x_tp, max_y_tp = uusimaa_municipality_buffer.total_bounds
# use extent to create dummy points and add them to list of coordinates
coords_tp = coords + [[min_x_tp, min_y_tp], [max_x_tp, min_y_tp],
[max_x_tp, max_y_tp], [min_x_tp, max_y_tp]]
# calculate Voronoi diagram
tp = Voronoi(coords_tp)# create empty list of hold Voronoi polygons
tp_poly_list = []
# create a polygon for each region
# 'regions' attribute provides a list of indices of the vertices (in the 'vertices' attribute) that make up the region
# Source: https://docs.scipy.org/doc/scipy/reference/generated/scipy.spatial.Voronoi.html
for region in tp.regions:
# Ignore region if -1 is in the list (based on documentation)
if -1 in region:
continue
else:
pass
if len(region) != 0:
# Create a polygon by using the region list to call the correct elements in the 'vertices' attribute
tp_poly_region = Polygon(list(tp.vertices[region]))
tp_poly_list.append(tp_poly_region)
else:
continue
# create GeoDataFrame from list of polygon regions
tp_polys = gpd.GeoDataFrame(tp_poly_list, columns=['geometry'], crs=uusimaa_municipality.crs)
# Clip polygon regions to the municipality boundary
tp_polys_clipped = gpd.clip(tp_polys, uusimaa_municipality)# Create subplots
fig, ax = plt.subplots(1, 1, figsize = (20, 20))
# Stylize plots
plt.style.use('bmh')
# Plot data
uusimaa_municipality.plot(ax = ax, color = 'none', edgecolor = 'dimgray')
tp_polys_clipped.plot(ax = ax, cmap = 'coolwarm', edgecolor = 'white', linewidth = 0.5)
uusimaa_cell.plot(ax = ax, marker = 'o', color = 'limegreen', markersize = 15)
# Set title
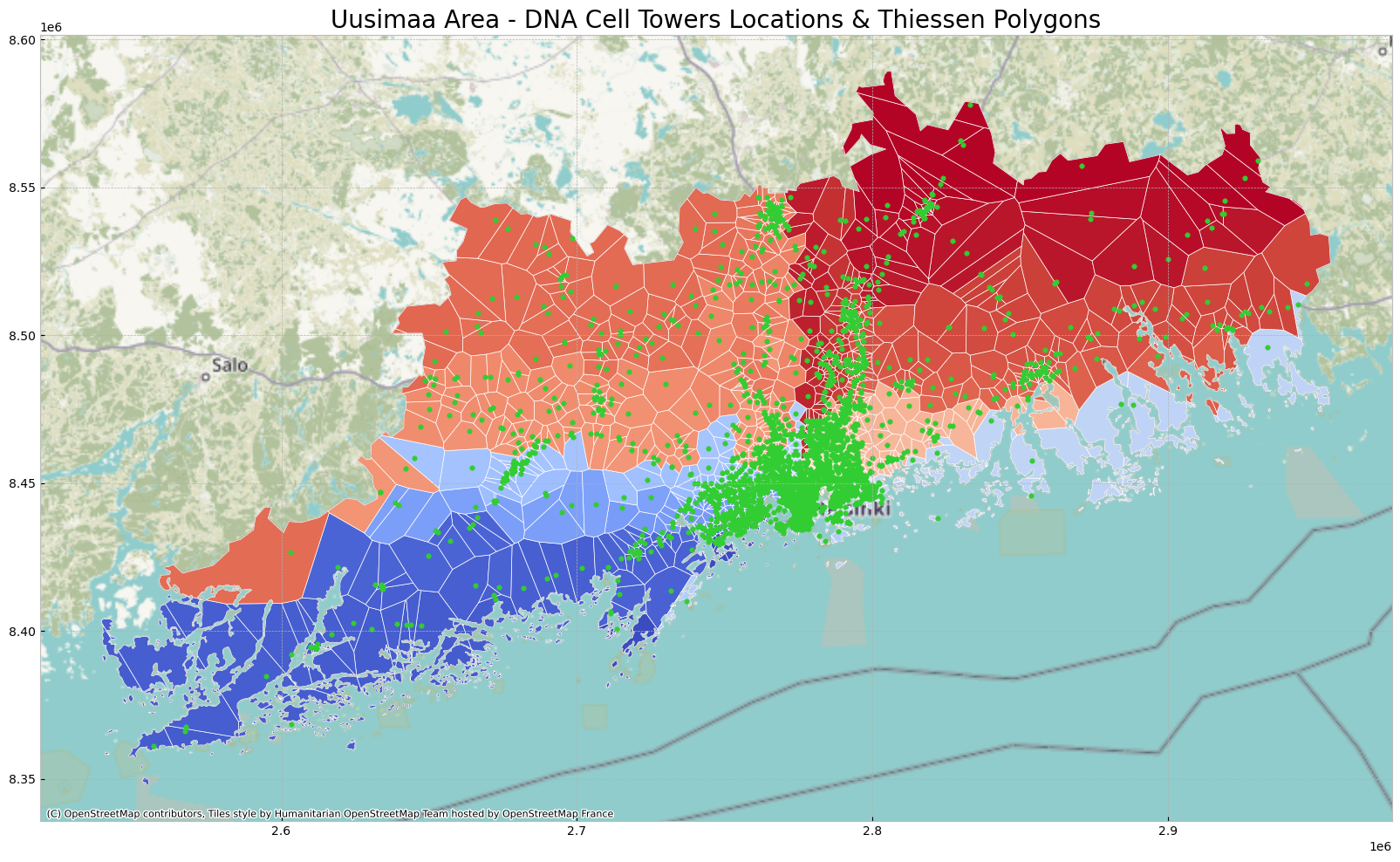
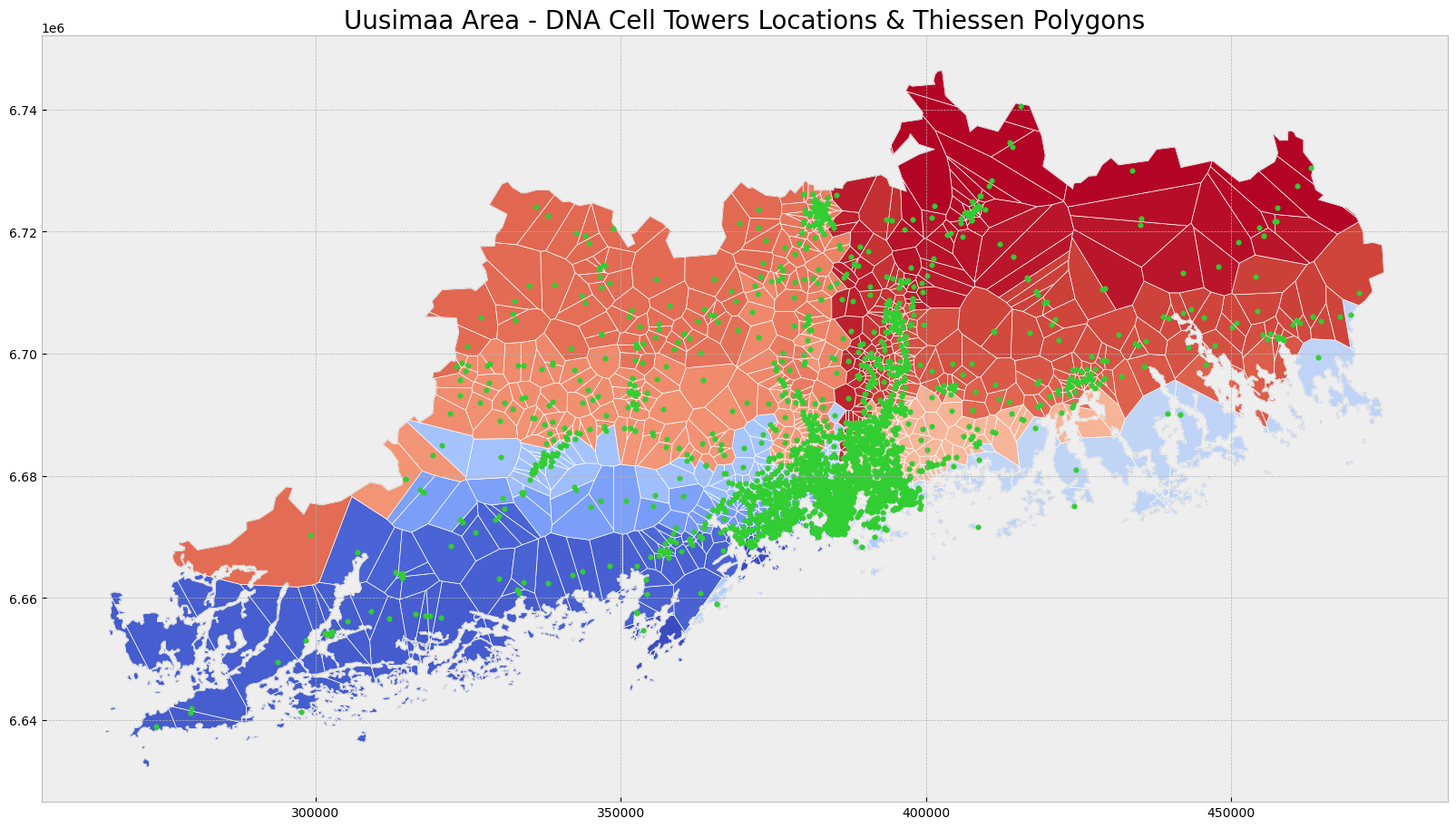
ax.set_title('Uusimaa Area - DNA Cell Towers Locations & Thiessen Polygons', fontdict = {'fontsize': '20', 'fontweight' : '3'})Text(0.5, 1.0, 'Uusimaa Area - DNA Cell Towers Locations & Thiessen Polygons')
# change coordinates system
uusimaa_municipality_new = uusimaa_municipality.to_crs(epsg=3857)
tp_polys_clipped_new = tp_polys_clipped.to_crs(epsg=3857)
uusimaa_cell_new = uusimaa_cell.to_crs(epsg=3857)
# plot data
ax_new = uusimaa_municipality_new.plot(figsize=(20,20), color = 'none', edgecolor = 'dimgray')
tp_polys_clipped_new.plot(ax = ax_new, cmap = 'coolwarm', edgecolor = 'white', linewidth = 0.5)
uusimaa_cell_new.plot(ax = ax_new, marker = 'o', color = 'limegreen', markersize = 15)
# add background tiles to plot
ctx.add_basemap(ax_new)
# Set title
ax_new.set_title('Uusimaa Area - DNA Cell Towers Locations & Thiessen Polygons', fontdict = {'fontsize': '20', 'fontweight' : '3'})Text(0.5, 1.0, 'Uusimaa Area - DNA Cell Towers Locations & Thiessen Polygons')